- Code: Select all
Goodness of Fit Results (TEST 2 + TEST 3) by Group
Group Chi-square df P-level
----- ---------- ---- -------
1 2.5084 9 0.9807
2 13.9081 10 0.1772
Total 16.4165 19 0.6293
I built a set of candidate Link-Barker models using PIM coding and sine links. Fletcher c-hat for my global model Phi(g*t) p(g*t) f(g*t) was 1.11.
The best-supported model was Phi(g) p(t) f(g). Since there was no time variation in Phi or f, I expected a single estimate of Phi, f, and Lambda for each group. Real parameter estimates looked as expected:
- Code: Select all
Parameter Estimate Standard Error Lower Upper
----------------- ----------- -------------- -------------- --------------
1:Phi 0.8063951 0.0298723 0.7411067 0.8583655
2:Phi 0.8885083 0.0211434 0.8398695 0.9237152
3:p 0.7539629 0.1089191 0.4922595 0.9064200
4:p 0.2471774 0.0414449 0.1750481 0.3368905
5:p 0.3905809 0.0456618 0.3055799 0.4827877
6:p 0.1943251 0.0341582 0.1358997 0.2700195
7:p 0.7183033 0.0513023 0.6080497 0.8073684
8:p 0.4941700 0.0525771 0.3927942 0.5960274
9:f 0.3146688 0.0552829 0.2174087 0.4314515
10:f 0.2351896 0.0495188 0.1520240 0.3453238
However, occasion 3 (the two-year interval) had substantially different derived estimates for Lambda:
- Code: Select all
Grp. Occ. Lambda-hat Standard Error Lower Upper
---- ---- -------------- -------------- -------------- --------------
1 1 1.1210639 0.0474039 1.0319363 1.2178893
1 2 1.1210639 0.0474039 1.0319363 1.2178893
1 3 1.2567842 0.1062857 1.0651281 1.4829263
1 4 1.1210639 0.0474039 1.0319363 1.2178893
1 5 1.1210639 0.0474039 1.0319363 1.2178893
2 1 1.1236979 0.0446491 1.0395386 1.2146706
2 2 1.1236979 0.0446491 1.0395386 1.2146706
2 3 1.2626970 0.1003442 1.0808388 1.4751539
2 4 1.1236979 0.0446491 1.0395386 1.2146706
2 5 1.1236979 0.0446491 1.0395386 1.2146706
When I fitted the equivalent model Phi(g) p(t) f(g) using the Pradel recruitment formulation, results were similar for Phi and p, but much different for f and Lambda:
- Code: Select all
Parameter Estimate Standard Error Lower Upper
------------------------- -------------- -------------- -------------- --------------
1:Phi 0.8032024 0.0300957 0.7375461 0.8556486
2:Phi 0.8866474 0.0213586 0.8375912 0.9222613
3:p 0.7023161 0.0822840 0.5217225 0.8361365
4:p 0.2535039 0.0400336 0.1832251 0.3395334
5:p 0.4428963 0.0495322 0.3491266 0.5409205
6:p 0.1692410 0.0296090 0.1188069 0.2353661
7:p 0.7001491 0.0513179 0.5911943 0.7903624
8:p 0.5158839 0.0532063 0.4124123 0.6180114
9:f 0.2026007 0.0362209 0.1406916 0.2827868
10:f 0.1454143 0.0286644 0.0976923 0.2109968
Grp. Occ. Lambda-hat Standard Error Lower Upper
---- ---- -------------- -------------- -------------- --------------
1 1 1.0058031 0.0326810 0.9437614 1.0719235
1 2 1.0058031 0.0326810 0.9437614 1.0719235
1 3 1.0116399 0.0657412 0.8907750 1.1489044
1 4 1.0058031 0.0326810 0.9437614 1.0719235
1 5 1.0058031 0.0326810 0.9437614 1.0719235
2 1 1.0320617 0.0277792 0.9790351 1.0879604
2 2 1.0320617 0.0277792 0.9790351 1.0879604
2 3 1.0651514 0.0573397 0.9585645 1.1835901
2 4 1.0320617 0.0277792 0.9790351 1.0879604
2 5 1.0320617 0.0277792 0.9790351 1.0879604
Since the unequal intervals seemed to be a problem, I edited the capture histories to have seven occasions with occasion 4 (2014) coded as zeroes. I then used the default one-year time intervals and fixed p = 0 for occasion 4. The estimates for both the Link-Barker and Pradel formulation were then exactly the same and there was no time variation in Lambda-hat:
- Code: Select all
Parameter Estimate Standard Error Lower Upper
------------------------ -------------- -------------- -------------- --------------
1:Phi 0.8032024 0.0286032 0.7411127 0.8533476
2:Phi 0.8866474 0.0202994 0.8403817 0.9207673
3:p 0.7023161 0.0782033 0.5312593 0.8308273
4:p 0.2535039 0.0380482 0.1863225 0.3349375
5:p 0.4428963 0.0470758 0.3535736 0.5360713
6:p 0.0000000 0.0000000 0.0000000 0.0000000 Fixed
7:p 0.1692410 0.0281406 0.1209667 0.2317023
8:p 0.7001491 0.0487729 0.5969241 0.7863984
9:p 0.5158839 0.0505677 0.4174393 0.6131111
10:f 0.2026007 0.0344246 0.1455579 0.2819981
11:f 0.1454143 0.0272429 0.1010434 0.2092697
Grp. Occ. Lambda-hat Standard Error Lower Upper
---- ---- -------------- -------------- -------------- --------------
1 1 1.0058031 0.0326810 0.9437613 1.0719234
1 2 1.0058031 0.0326810 0.9437613 1.0719234
1 3 1.0058031 0.0326810 0.9437613 1.0719234
1 4 1.0058031 0.0326810 0.9437613 1.0719234
1 5 1.0058031 0.0326810 0.9437613 1.0719234
1 6 1.0058031 0.0326810 0.9437613 1.0719234
2 1 1.0320617 0.0277792 0.9790351 1.0879604
2 2 1.0320617 0.0277792 0.9790351 1.0879604
2 3 1.0320617 0.0277792 0.9790351 1.0879604
2 4 1.0320617 0.0277792 0.9790351 1.0879604
2 5 1.0320617 0.0277792 0.9790351 1.0879604
2 6 1.0320617 0.0277792 0.9790351 1.0879604
I assume these estimates are correct, but I don't understand why the unequal intervals made such a difference for the Link-Barker formulation. My understanding from the MARK book was that either method of coding the capture histories should have given the same results.
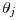 to the interval-specific value
to the interval-specific value 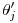 uses
uses 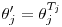 where
where 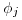 or
or 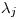 , and
, and 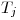 is the duration of interval
is the duration of interval 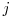 .
.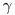 follows the same pattern except that the relevant duration for
follows the same pattern except that the relevant duration for  is
is  . Scaling per capita recruitment
. Scaling per capita recruitment 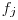 is more tricky. We use
is more tricky. We use 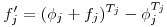 .
.