Here is an example with the dipper data using made up rain levels instead of flooding. I create a model in which rain is a design covariate and then I create 6 individual covariates specifically named r1,r2,...r6 to match the time values of 1 to 6. r1 differs from say r2 but all the values of r1 are the same. Evan has used the same example in the sidebar at the end of section 11.5 to show how the same thing can be done with the MARK interface. You get the same estimates if you treat the covariates as design or individual covariates but using as an individual covariates let's you get predictions easily.
- Code: Select all
library(RMark)
data(dipper)
# add a time varying covariate for Phi named to match beginning time of each time interval (default begin.time=1 and time.intervals=1)(r1,r2,..r6)
dipper$r1=rep(1,294)
dipper$r2=rep(10,294)
dipper$r3=rep(8,294)
dipper$r4=rep(15,294)
dipper$r5=rep(3,294)
dipper$r6=rep(6,294)
# process data
dp=process.data(dipper)
# create default design data
ddl=make.design.data(dp)
# add rain environmental covariate to design data for Phi; this matches r1 to r6.
ddl$Phi$rain=1
ddl$Phi$rain[ddl$Phi$time==2]=10
ddl$Phi$rain[ddl$Phi$time==3]=8
ddl$Phi$rain[ddl$Phi$time==4]=15
ddl$Phi$rain[ddl$Phi$time==5]=3
ddl$Phi$rain[ddl$Phi$time==6]=6
# fit model using design covariate
modcov=mark(dp,ddl,model.parameters=list(Phi=list(formula=~rain)))
# fit model using individual covariate
modicov=mark(dp,ddl,model.parameters=list(Phi=list(formula=~r)))
# get and plot predictions using individual covariates
predictions=covariate.predictions(modicov,data=data.frame(r1=0:20),indices=1)$estimates
with(predictions,
{
plot(0:20,estimate,xlab="Rain",ylab="Survival",ylim=c(0,1))
lines(0:20,lcl,lty=2)
lines(0:20,ucl,lty=2)
})
![$\widehat{\mbox{var}}\bigl(\hat{\bar{p}}\bigr)=\sum_{i=1}^R{w}_i\Bigl[\widehat{\mbox{var}}\bigl(\hat{p}_i\bigm| M_i\bigr)+\bigl(\hat{p}_i-\hat{\bar{p}}\bigr)^2\Bigr]$ $\widehat{\mbox{var}}\bigl(\hat{\bar{p}}\bigr)=\sum_{i=1}^R{w}_i\Bigl[\widehat{\mbox{var}}\bigl(\hat{p}_i\bigm| M_i\bigr)+\bigl(\hat{p}_i-\hat{\bar{p}}\bigr)^2\Bigr]$](/forum/latexrender/pictures/36654d196cdc64473e36a5564e428027.gif)
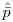 . If you do the averaging on the logit scale, then back-transform, you get a different average (and thus, different estimates of the unconditional variance) than if you back-transform the estimates for each model first, and then average -- and do subsequent calculations -- on the real scale. Jensen's inequality.
. If you do the averaging on the logit scale, then back-transform, you get a different average (and thus, different estimates of the unconditional variance) than if you back-transform the estimates for each model first, and then average -- and do subsequent calculations -- on the real scale. Jensen's inequality. ![$\widehat{\mbox{var}}\bigl(\mbox{logit}\hat{\bar{p}}\bigr)=\sum_{i=1}^R{w}_i\Bigl[\widehat{\mbox{var}}\bigl(\mbox{logit}~\hat{p}_i\bigm| M_i\bigr)+\bigl(\mbox{logit}~\hat{p}_i-\mbox{logit}\hat{\bar{p}}\bigr)^2\Bigr]$ $\widehat{\mbox{var}}\bigl(\mbox{logit}\hat{\bar{p}}\bigr)=\sum_{i=1}^R{w}_i\Bigl[\widehat{\mbox{var}}\bigl(\mbox{logit}~\hat{p}_i\bigm| M_i\bigr)+\bigl(\mbox{logit}~\hat{p}_i-\mbox{logit}\hat{\bar{p}}\bigr)^2\Bigr]$](/forum/latexrender/pictures/16bb9b38da6659efab488e39af10cb1e.gif)